Genomics and Physiopathology of Neurodevelopmental & Motor Neuron Disorders
Leader: Dr. Frédéric Laumonnier
Co-leader: Pr. Patrick Vourc’h
Co-leader: Pr. Patrick Vourc’h

The identification of specific and sensitive markers of brain diseases has become a major issue in public health and a priority in biomedical research. The general objective of our Team is to identify and characterize clinical, molecular (i.e. genetic, epigenetic, biochemical) and pathophysiological markers in neurodevelopmental disorders (NDD) and Motor Neuron Diseases (MND), one of the major challenges in neuropsychiatry and in neuroscience. These simplex or multiplex markers would allow early management of patients, as well as personalized predictive and participatory medicine. Our project is rooted in a translational research approach: integration of clinical and biological data, DNA sequencing combined with a detailed characterization of the impact of identified gene mutations on neuronal development and aging, and on the brain's structural and functional connectivity in pathophysiological models (cellular, animal).
Our research is strongly integrated into a rich environment with membership and/or coordination of national and international networks, such as 3 Reference centers for rare diseases within the Tours University Hospital, the Labex MabImprove, FHU GenoMeds, FILSAN (on ALS), SFEIM (inborn errors of metabolism), MinE (ALS), GOLD (Cross-cutting program on genomic variability, Inserm) programs. Our scientific approach benefits from the multidisciplinary dimension of the team (geneticists, neurologists, neurobiologists, biochemists, molecular biologists, ophtalmologists) and from strong academic and industrial collaborations.
Objectives
To identify and characterize clinical, molecular and pathophysiological markers in Intellectual Disability (ID) and in Amyotrophic Lateral Sclerosis (ALS)
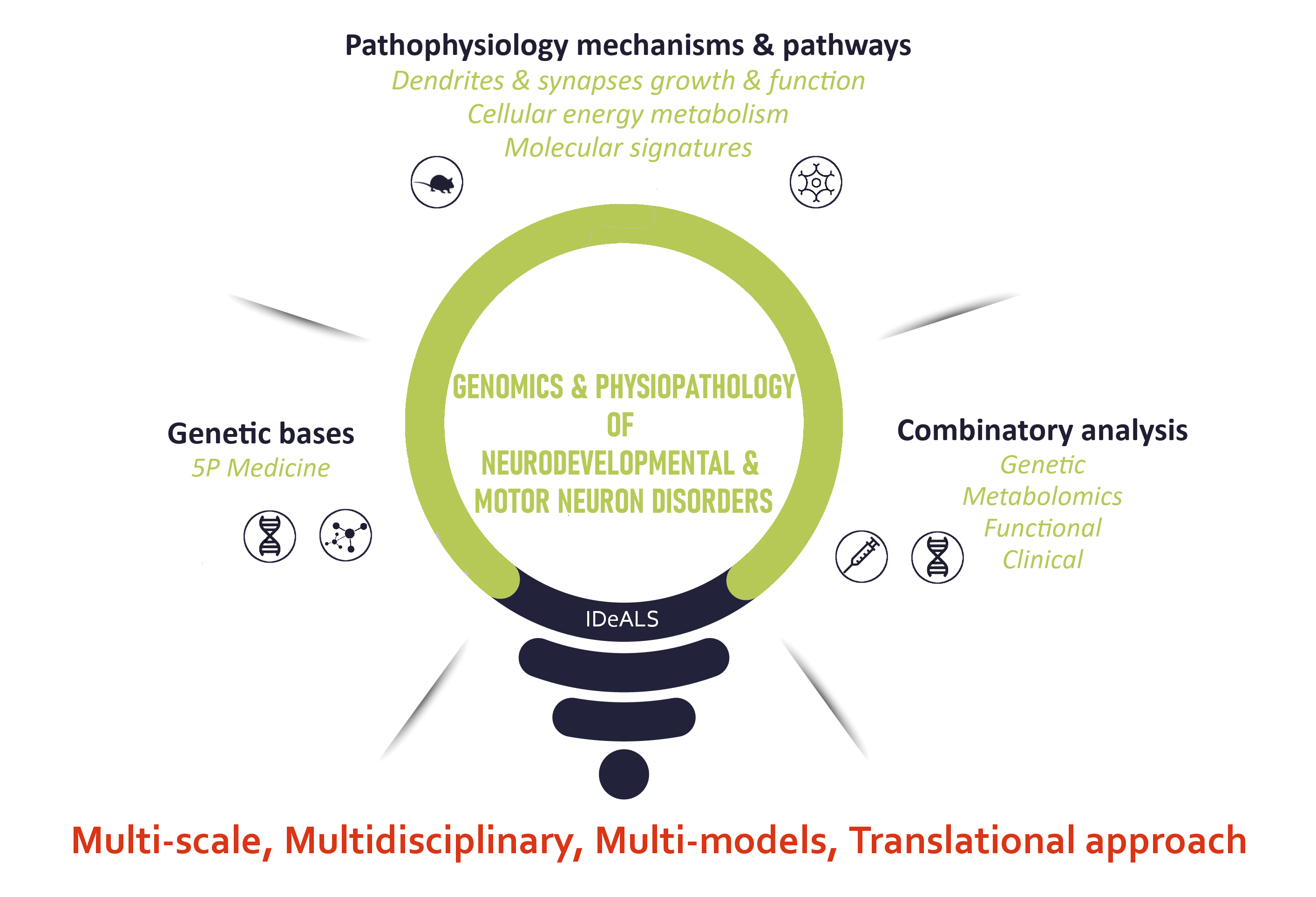
The team’s project is organized around three main axes: (1) the genetic bases of NDD (intellectual disability) and MND (amyotrophic lateral sclerosis), (2) translational approaches to investigate pathophysiological mechanisms, and (3) integration of data for clinical transfer.
Specific research axes
Improving knowledge on the genetic bases of ID and ALS
This axis aims to identify new simplex or multiplex disease markers using clinical, biological and molecular (genetics, epigenetics, genomics, epigenomics, metabolomics) approaches to allow earlier and more effective management of patients, and to develop 5P medicine (predictive, preventive, personalized, participatory, and purpose-driven approach). Our research activity on the genetic bases of NDD and ALS is directly linked to the Departments of Genetics, of Neurology and of Biochemistry and Molecular biology of the University Hospital of Tours which are labelled Reference Laboratories in Medical Biology (LBMR) on ID and ALS.Deciphering pathophysiological mechanisms through translational strategies
We aim to better understand the pathophysiological mechanisms involved in ID and ALS using both global and targeted approaches on previously identified markers. We focus on alterations in signaling pathways involved in the growth and function of dendrites and synapses, and on perturbations in cellular energy metabolism. In addition, we aim to establish molecular signatures linked to genetic alterations. We use both in vitro and in vivo approaches, using primary neuronal cultures, fibroblast cultures, human iPSCs, and mouse models. A particular innovative aspect of this axis is the integrated multi-omics approach available in the team and in the Unit at tissue and cell levels (single cell RNA sequencing, metabolomics), and to linking them to clinical-biological data.Epigenetic mechanisms regulating brain-expressed genes and potentially involved in brain disorders
We identified a primate specific epigenetic factor, SETMAR (histone methyl transferase), as being involved in human fetal brain development. Some of SETMAR target genes are involved in biological pathways that are of interest for our team. The actual involvement of SETMAR in these mechanisms is evaluated and the epigenetic regulation (related or not to SETMAR) of the team's main candidate genes is investigated. We also explore the impact of exposure of environmental toxins leading to neuropathic consequences, by identifying genes whose dysregulation disrupts stem cell differentiation in two complementary models: human Glioblastoma (GBM) stem cells and neural stem cells.Combinatory and integrative analyses for preclinical and clinical transfer
Genetic, metabolomic, functional and clinical data are combined to improve the diagnostic assessment of developmental disorders and motor neurons diseases. This axis is part of a research framework ranging from preclinical research to clinical trials (ALS, Phenylketonuria - PKU). We also use data from our works on pathophysiological mechanisms to design and test new therapeutic strategies, via intrabodies approaches (team member of Labex MabImprove) or small chemical molecules.
Functional groups
Genetic architecture and pathophysiology of neurodevelopmental disorders
Resp.: Dr. Frédéric Laumonnier
Objectives: 
Objectives:
- To improve knowledge of the genetic architecture of NDD
- To resolve diagnostic deadlocks through functional analysis of genetic variants
- To explore pathophysiological mechanisms involved in NDD using neuronal and animal models

Several projects are currently underway:
- ASDecode (ANR): Translational research targeting the PTCHD1 pathway involved in NDD
- IDGePhe-PAK (ANR): Analysis of the genotype-phenotype correlation in ID to improve diagnosis and propose personalized medicine: PAK3 as a representative example
- UPS-NDDecipher (ANR): Translational investigation of NDD caused by impairment of the Ubiquitin-Proteasome System
- UPS-NDDiag (EJP-RD ERare): Development of diagnostic solutions for NDD caused by ubiquitin-proteasome system dysfunction
Exploration of Amyotrophic Lateral Sclerosis (ALS)
Resp.: Pr. Patrick Vourc'h
Objectives: 
Several projects are currently underway: 
Objectives:
- To better understand the etiology of ALS and identify multiplex biomarkers using clinical, genetic and metabolomic approaches
- To characterize pathophysiological mechanisms using in vitro and in vivo approaches

Several projects are currently underway:
- FG-Coals (ANR): French-German cohort study to determine factors associated with weight loss in amyotrophic lateral sclerosis
- PremodiALS (JPND): Linking pre-diagnosis disturbances of physiological systems to Neurodegenerative Disease

Metabolomics and new therapeutic strategies
Resp.: Pr. Patrick Emond
Objectives:  Several projects are currently underway:
Several projects are currently underway:

Objectives:
- To develop innovative metabolomics approaches for studies of diseases of the central nervous system
- To characterize neurocognitive or motor functions of mouse models of ID, ALS and PKU
- To design new therapeutic strategies in ALS focusing in particular on protein aggregates or altered molecular pathways
- To identify Alzheimer’s disease biomarkers in tears

- PeliCo (ANR): Towards a new therapeutic strategy targeting cytoskeleton dynamics by developing stapled Peptides to selectively inhibit LIMKs/Cofilin interaction (ALS)

Epigenetic Modulation of Neuronal Development and Glioblastoma Biogenesis: Impacts of SETMAR and Environmental Pollutants
Resp.: Pr. Corinne Augé
Objectives:
Objectives:
- To explore the role of SETMAR, a human specific epigenetics factor, in neurons and astrocytes differentiation
- To understand the protective role of SETMAR against glioblastoma
- To evaluate the impacts of environmental pollutants on neurons and astrocytes differentiation and on glioblastoma stem cells
- ANR-preciput (University of Tours) : SETMAR mRNA Sonoporation impact on neuronal stem cells
- Ligue contre le Cancer : SETMAR mRNA Sonoporation impact on Glioblastoma Stem cells (SONOSET)
- APR-IA Région Centre Val de Loire. Watershed of Loire basin, pollutants and neurodevelopment (VECTRA)

French presentation
Présence d'un lecteur vidéo
Fundings










Industrial partners


Medias
Présence d'un lecteur vidéo